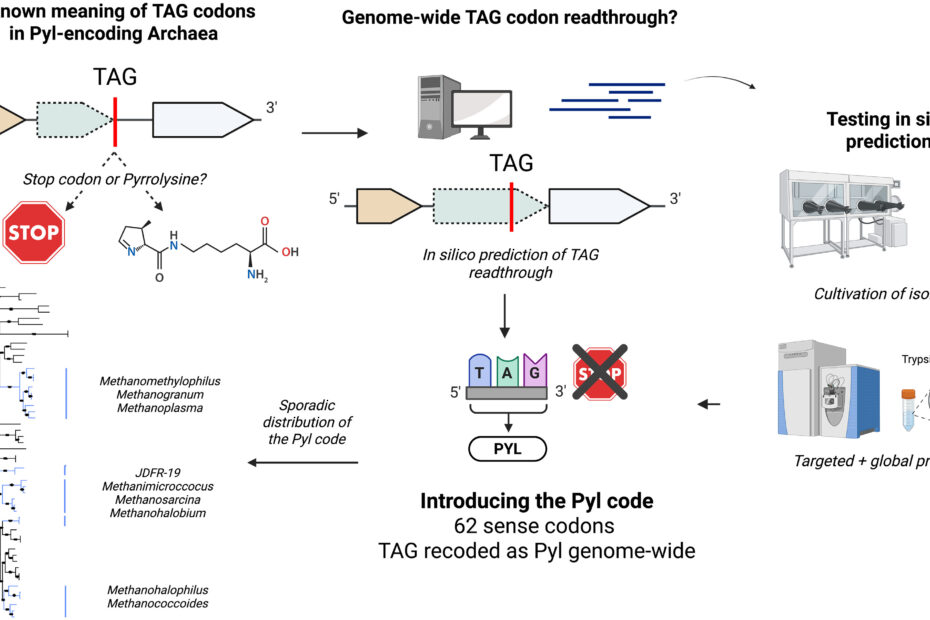
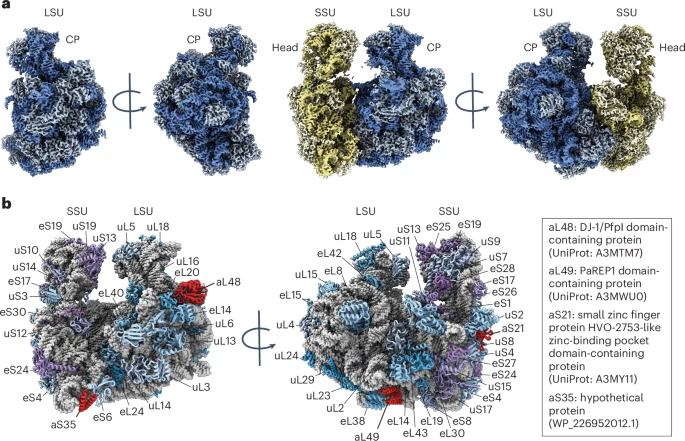
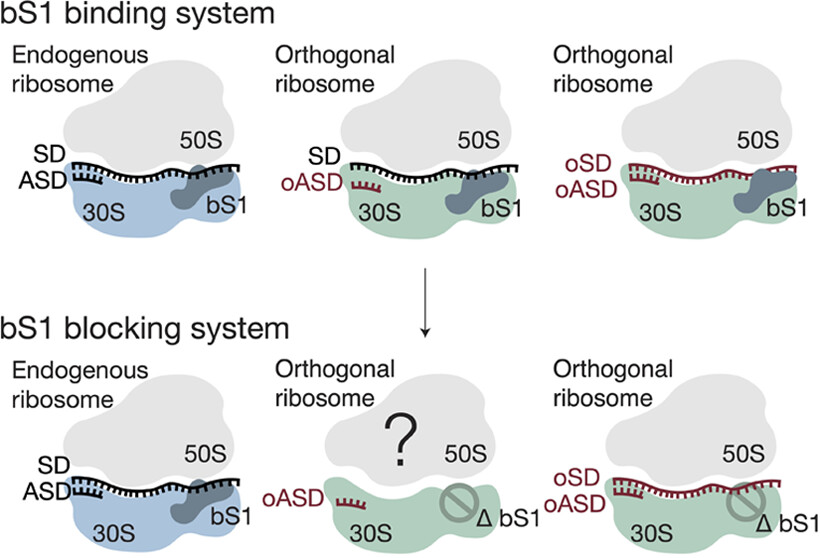
An archaeal genetic code with all TAG codons as pyrrolysine. (Science, 2025)
Many genetic code expansion (GCE) techniques rely on the native ability that some organisms have to recode the stop codon TAG to incorporate pyrrolysine, a special amino acid not encoded in other organisms. Previously, in known organisms with this ability, pyrrolysine was only incorporated at… Read More »An archaeal genetic code with all TAG codons as pyrrolysine. (Science, 2025)