Yuna Lee (left) and Emma Yin (right) are undergraduate students who participated in the 2021 C-GEM summer research program. They are working to create a Pyrrolysine synthetase (PylRS) database, which compiles information on diverse, non-canonical amino acid substrates for the wild type enzyme as well as evolved PylRS derivatives. The database aims to increase accessibility for researchers using genetic code expansion to develop the next generation of nanomaterials and pharmaceutical drugs.
Introduction
There are 20 canonical amino acids present in all biological systems, providing a limited functionality for the production of proteins.1 In an effort to create proteins that have unique biochemical properties, engineered aminoacyl tRNA synthetases- tRNA pairs may be used to encode non-canonical amino acids in living cells.1
Together with multiple other undergraduate students from various universities, we are working to create a database of information compiling tools used for genetic code expansion. The goal of the Center for Genetically Encoded Materials (C-GEM) is to repurpose translational machinery to create new chemical matter. Throughout the 2021 summer undergraduate research program, our team focused on developing a database of pyrrolysyl-tRNA synthetase (PylRS) information. PylRS encodes pyrrolysine, the 22nd proteinogenic amino acid discovered in Methanosarcina barkeri.1 The structure of PylRS has a large hydrophobic side chain-binding pocket for the binding of pyrrolysine. PylRS displays a uniquely high tolerance toward variations of the substrate side chain, low selectivity toward the substrate ɑ-amine, and low selectivity toward its tRNA anticodon.1
PylRS and tRNAPyl can be extensively used to add non-canonical amino acids to the genetic code of bacterial and eukaryotic cells.1 Typically, PylRS enzymes from Methanosarcina mazei (Mm) or Methanosarcina barkeri (Mb) are used to add non-canonical amino acids using genetic code expansion.1 Non-canonical amino acids can be described as any amino acids that deviate from the original 20, and they contain many chemically unique reactive groups that allow for the installation of a large variety of biochemical and biophysical probes for protein function investigation and biotechnological development.1 Due to its high tolerance for different substrates, PylRS is commonly used to covalently link noncanonical amino acids onto tRNAs.1 This PylRS incorporation system has allowed access to proteins with multiple posttranslational modifications mainly on lysine both in vitro and in vivo, making it possible to show how these modifications regulate protein functions, enzyme activities, and nucleosome interactions with transcription factors.1
The creation of our PylRS database specifically focuses on the uses of native and mutated PylRS for the incorporation of non-canonical amino acids into proteins. Our database will provide a searchable resource to other researchers in the genetic code expansion field that will help support diverse monomer incorporation efforts. As we progress through the summer, our knowledge of specific biochemical themes broadens and helps us to further understand the complexities of synthetases. There are many aspects about PylRS and synthetases that we are continuing to discover, but as we set up this database and input further data regarding PylRS, we can ensure that the field of ribosomal research will benefit from the accessibility of synthetase information.
Methods
To inform our compilation of relevant information regarding aminoacyl tRNA synthetases into a database, we read numerous research papers about PylRS mutants and their functions. Our database includes information on mutations, non-canonical amino acid monomer structures, kinetic parameters, and tRNA types. Our main source for finding relevant research papers is the review paper titled “Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool”, which helped us form a foundation of understanding regarding PylRS so that we could understand and dissect the various research articles henceforth.1 Many of the citations used by this review paper were read and put into the database, as well as other research articles from Nature, Nature Chemistry, and ACS Chemical Biology. The molecule editor program, ChemDraw, was similarly utilized to draw out the non-canonical monomer structures provided in the database.
Results
The goal in creating a database that compiles specific information about engineered PylRS and the non-canonical amino acids that were incorporated into proteins is to provide a reliable collection of information for researchers to utilize in future research or experiments. As genetic code expansion has been around for about 20 years, many noncanonical amino acids have been incorporated using orthogonal synthetases, each with various applications and uses.1 However, there is no searchable resource of the amino acids and engineered synthetases discovered. By creating a database where scientists can have access to centralized information regarding PylRS data, researchers may be better able to utilize this information in related experiments.
Discussion

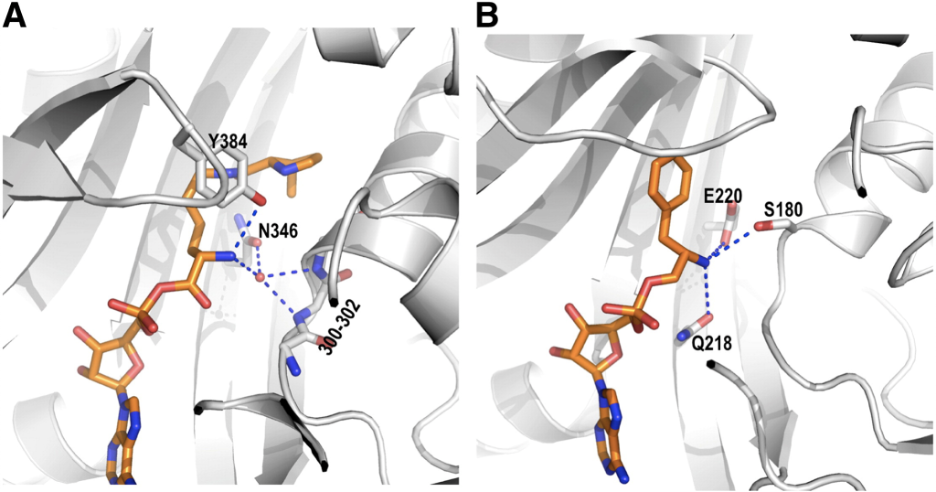
The primary function of aminoacyl tRNA synthetases is to catalyze the aminoacylation of transfer RNAs (tRNAs).2 PylRS variants and their cognate tRNAPyl are specifically utilized to add non-canonical amino acids to the genetic code of prokaryotic and eukaryotic cells at amber stop codons.1 Due to low evolutionary selection pressure, PylRS has uniquely high substrate chain promiscuity allowing for recognition of many lysine-like hydrophobic side chains (Fig.1). Additionally, PylRs show low selectivity toward the tRNA anticodon, which allows for customization of desired codon suppression. Finally, PylRS also shows low selectivity toward the ɑ-amine on amino acids by coordinating a water molecule to the α-amine instead of directly hydrogen bonding the α-amine like other synthetases (Fig. 2). This may increase the scope of genetic code expansion by allowing novel substitutions in the ɑ-amine position.

To date, many of the non-canonical amino acids charged by PylRS have the unique characteristic of bioorthogonal reactive handles included in their structure (Fig. 3).3 The chemical functional groups on these amino acids do not react with biological molecules but can still undergo specific reactions, such as the selective attachment of dyes to non-canonical amino acids, allowing researchers to visualize specific proteins.2 Bioorthogonal reactive handles also allow for coupling to other proteins, as well as modifying proteins to make more stable protein therapeutics.2
Techniques for the genetic encoding of new amino acids into native cell-types of neuroendocrine, muscle, neurons and live animals are especially exciting, as they will permit the use of encoded reporters for optogenetic applications as well as having the promise to yield transformative insights into molecular physiology and the basis for disease.3 Essentially, PylRS and its derivative can allow for the synthesis of proteins with novel characteristics, accounting for numerous potential applications.
Creating a database of information that can help further research on synthesizing chemically diverse polymers by the ribosome has allowed us to learn about synthetases and, generally, the world of research. Novel materials may improve the longevity of synthesized biomimetic materials and provide better properties to be used in research across many different fields.2 Over the course of the summer undergraduate research program with C-GEM, we focused our attention on the research that has been done on PylRS, while future students may focus on other engineered synthetases. In conclusion, the work CGEM is doing has inspired us to think outside the box and pursue areas of science that have rarely been explored before.
References
1. Wan, W.; Tharp, J. M.; Liu, W. R. Pyrrolysyl-TRNA Synthetase: An Ordinary Enzyme but an Outstanding Genetic Code Expansion Tool. Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics 2014, 1844 (6), 1059–1070. https://doi.org/10.1016/j.bbapap.2014.03.002.
2. Ovaa, H.; Wals, K. Unnatural Amino Acid Incorporation in E. Coli: Current and Future Applications in the Design of Therapeutic Proteins. Front. Chem. 2014, 2. https://doi.org/10.3389/fchem.2014.00015.
3. Leisle, L.; Valiyaveetil, F.; Mehl, R. A.; Ahern, C. A. Incorporation of Non-Canonical Amino Acids. Adv Exp Med Biol 2015, 869, 119–151. https://doi.org/10.1007/978-1-4939-2845-3_7.
