Alexis Vanzandt (left) and Kaitlyn Szalay (right) are undergraduate students who participated in the C-GEM summer research program. Their project focused on analyzing and integrating antibiotic-ribosome mutation data to aid in the accessibility of ribosome information and propel future studies.
Introduction
During the Center for Genetically Encoded Materials (C-GEM) summer undergraduate research program, we compiled information on antibiotics that target the ribosome and ribosomal residues that are involved in antibiotic activity and antibiotic resistance. Both of us were interested in seeing research first-hand and exploring different career options during our undergraduate before deciding on a path to take. As part of team ribosome, we read and analyzed publications that discuss mutations involving antibiotics, nucleotides, and other molecules that affect the ribosome and have begun pulling out key information to help build a database.
This database will end up being a central pool of information about engineered translation systems that will help C-GEM and other research groups re-engineer the cellular translation process to incorporate chemically diverse monomers. This has great potential for the future to advance science and technology in different areas including the environment and health. New medications can be designed, more stable vaccines that do not need to be refrigerated, and creation of new polymers that are biodegradable are just some of the possibilities with an expanded set of monomers.
Cataloging antibiotic binding sites or mutations of the ribosome that affect antibiotic resistance are important because the bacterial ribosome is a critical target for antibiotics. The job of the ribosome is to synthesize all of the proteins in the cell. If this function is blocked due to antibiotics, the cell is not able to continue to grow and survive. Bacterial ribosomes are different enough from eukaryotic ribosomes so the antibiotics will act on the bacteria and not affect our own cells. Drugs can inhibit or enhance ribosomal activity by interfering with messenger RNA (mRNA) or other RNA binding proteins and these effects are important to know about before starting an experiment.
When mutations occur in the ribosome, the ability for antibiotics to bind to the ribosome can be affected as the mutations occur in active sites or near the binding sites and these are typically deleterious mutations that affect antibiotic binding.1,3,4,5, Deleterious mutations are mutations that increase susceptibility to certain disorders. With the mutations in the ribosome, it has been seen that the ribosome has greater resistance to certain antibiotics especially those that bind to the ribosomal peptidyl transferase center (PTC) in the large subunit of the ribosome. One such example is the L3 (Fig. 1A) protein near the surface of the 50S (Fig. 1B) subunit that has a loop close to the PTC.3 Mutations in the L3 loop near the PTC are associated with increased bacterial resistance to antibiotics such as tiamulin.3 During the time that an antibiotic binds to a specific area/subunit within the ribosome, other outcomes are shown.
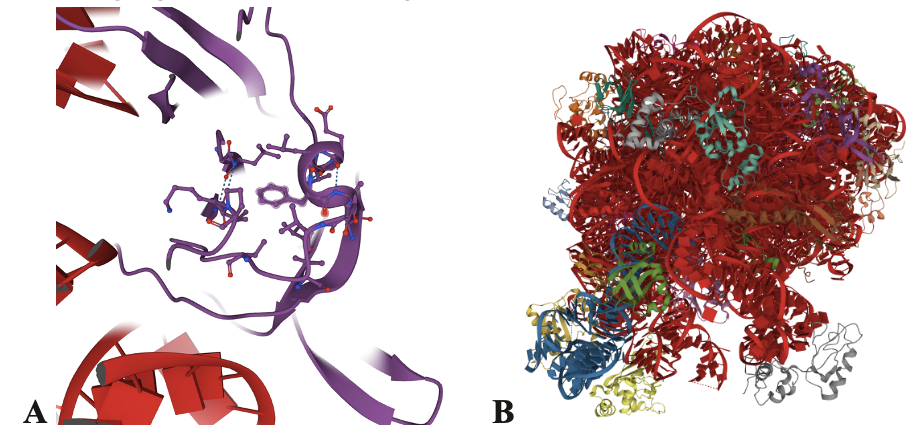
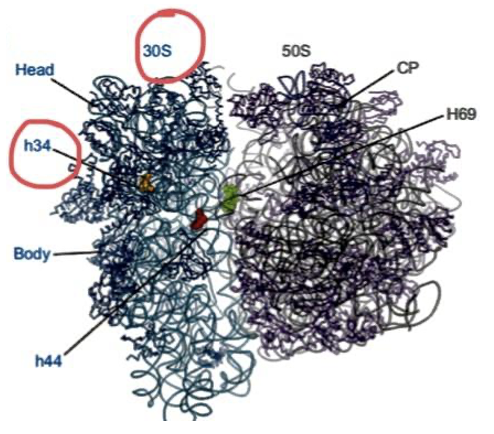
We’ve learned from scientific reviews that antibiotics can also induce conformational changes within the ribosome. Once the antibiotic binds it can inhibit or enhance ribosomal activity. For example, antibiotic binding can change the ribosome’s conformation, which is its spatial arrangement, affecting the particular subunit in which the antibiotic is bound or enzyme activity within the ribosome. In the case of spectinomycin (Fig. 3), high resolution structures and biochemical data showed that binding to the h34 region of the small subunit causes a conformational change that reduces ribosome activity and mRNA translation.2 This paper allowed a better understanding of the antibiotic-ribosome relationship because it used high resolution structures to generate hypotheses that were further investigated using biochemical assays.
Key words
Ribosome: A molecular machine within a cell that functions to synthesize proteins.
RNA: (Ribonucleic acid) is a polymeric molecule used in various biological roles in coding, decoding, regulation and expression of genes.
mRNA: Messenger RNA (mRNA) carries genetic information copied from DNA in a three-base code, which each specify a particular amino acid
tRNA: Transfer RNA (tRNA) is a molecule composed of RNA that carries amino acids to the ribosome to be added to a chain of amino acids.
Amino acid: The building blocks of proteins. It is an organic compound that contains one at least one amino group and one carboxylic acid group attached to a central carbon. This carbon is known as the alpha with which a variable side chain is bound.
50S subunit: in prokaryotes, this is the large subunit of the 70S ribosome that contains 23S ribosomal RNA (rRNA), 5S rRNA, and proteins L1-L77
30S subunit: in prokaryotes, this is the small subunit of the 70S ribosome that contains 16S ribosomal RNA (rRNA) and 21 proteins in E. coli.
rRNA: ribosomal RNA (rRNA) comprises part of the ribosome and is responsible for its catalytic activity.
PTC: the ribosomal peptidyl transferase center in the large subunit of the ribosome.
Mutation: A change in genetic sequence. (Ex: the substitution of a nucleotide base with another nucleotide base)
Biopolymer: natural polymers produced by the cells of living organisms.
Antibiotics: chemical compounds used to kill or inhibit the growth of bacteria.
Non-canonical amino acids: amino acids incorporated into the growing polypeptide chain as proteins are being synthesized at the ribosome. These are unnatural amino acids because they are typically not found in natural polypeptide chains. Polymer: A group of monomers joined together.
Methods
The papers and reviews have come from library database searches and mentor recommendations. We have also come across papers or reviews by looking at the citations of papers we read or seeing what papers have cited papers we already read. As we analyze and deconstruct the reviews and papers, we pull key information and data about specific antibiotic-ribosome mutations. This data includes information about how the antibiotic or ribosome is affected once binding occurs, nucleotides the antibiotic comes into contact with, and the experiments and methods the authors of the paper used to conduct their research. These data provide insight on how antibiotics affect the ribosome, which residues are involved, and how ribosomal mutations impact antibiotic action. These effects range depending on the antibiotics and mutations that occur.
Results
While analyzing and deconstructing these peer-reviewed papers we were able to make specific conclusions. We noticed patterns and trends that occurred once an antibiotic bound to the ribosome. For example, mutations, especially near prominent binding sites, will cause the bacteria to become resistant to antibiotics.1,3,4,5 By our observation, most antibiotics that bind to sites within the smaller subunit (30S), including spectinomycin, tend to induce a conformational change that disrupts translation.2 We also noticed that antibiotics which bind to the large subunit usually target the PTC. Mutations within this region tend to cause bacterial resistance to antibiotics. We also discovered that the antibiotics could come into contact with specific residues near their target site of the ribosome.
Discussion
There are many potential implications this research will have for the future. Knowing how antibiotics can affect the ribosome or how mutations in the ribosome affect the bacteria’s susceptibility to different antibiotics will be useful to researchers who use antibiotics as tools for basic research. It will also be helpful when designing drugs that bind to the ribosome and to see how it will affect the ribosome once bound. The information in the database can also be utilized by ribosome engineers interested in which regions of the ribosome are tolerant to mutations and how these mutations might affect antibiotic sensitivity. Repurposing the ribosome to accept exotic monomers is the overall goal and it will affect many areas of research in medicine, environment, health. Some future ideas that can be explored with engineered ribosomes include generating biopolymers capable of harvesting solar energy, making plastic alternatives that are biodegradable, stable vaccines that do not need to be refrigerated, as well as having bacteria that can create therapeutics when needed making it more affordable.
References
1. Argudín, M. A.; Roisin, S.; Dodémont, M.; Nonhoff, C.; Deplano, A.; Denis, O. Mutations at the Ribosomal S10 Gene in Clinical Strains of Staphylococcus Aureus with Reduced Susceptibility to Tigecycline. Antimicrob Agents Chemother 2018, 62 (1). https://doi.org/10.1128/AAC.01852-17.
2. Borovinskaya, M. A.; Shoji, S.; Holton, J. M.; Fredrick, K.; Cate, J. H. D. A Steric Block in Translation Caused by the Antibiotic Spectinomycin. ACS Chem. Biol. 2007, 2 (8), 545–552. https://doi.org/10.1021/cb700100n.
3. Cocozaki, A. I.; Altman, R. B.; Huang, J.; Buurman, E. T.; Kazmirski, S. L.; Doig, P.; Prince, D. B.; Blanchard, S. C.; Cate, J. H. D.; Ferguson, A. D. Resistance Mutations Generate Divergent Antibiotic Susceptibility Profiles against Translation Inhibitors. Proc Natl Acad Sci USA 2016, 113 (29), 8188–8193. https://doi.org/10.1073/pnas.1605127113.
4. Klitgaard, R. N.; Ntokou, E.; Nørgaard, K.; Biltoft, D.; Hansen, L. H.; Trædholm, N. M.; Kongsted, J.; Vester, B. Mutations in the Bacterial Ribosomal Protein L3 and Their Association with Antibiotic Resistance. Antimicrob. Agents Chemother. 2015, 59 (6), 3518–3528. https://doi.org/10.1128/AAC.00179-15.
5. Vester, B.; Douthwaite, S. Macrolide Resistance Conferred by Base Substitutions in 23S RRNA. Antimicrob Agents Chemother 2001, 45 (1), 1–12. https://doi.org/10.1128/AAC.45.1.1-12.2001.
6. Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S. K.; Koča, J.; Rose, A. S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Research 2021, 49 (W1), W431–W437. https://doi.org/10.1093/nar/gkab314.