Dina Tekle (left) and Nicole Rivera (right) are undergraduate students in the 2021 C-GEM summer research program, who are integrating ribosomal mutation data for the purpose of accelerating research via increasing the accessibility of ribosome information.
Introduction
Imagine a world with cleaner air, less waste and clean water that is easily accessible to everyone. A world where the latest advances in biochemistry can deliver a drug to specific targets within the human body, vastly expanding the limits of modern medicine. A world where basic life-saving medication can be made more readily available to more people with advanced shelf stabilization techniques. The center for Genetically Encoded Material’s (C-GEM) mission is to aid in the creation of such a world by modifying the ribosome (nature’s protein synthesizing machine) to create specific molecules that can be linked together to create novel polymers.1 These polymers can then be modified to create things like: biodegradable plastics, nanoparticles and bioconjugates that can deliver medication to previously unreachable areas within the human body, new and more efficient biofuels, new methods for attaining sustainable clean water, as well as endless other possibilities. Our work for the summer undergraduate research program involves reading research literature on ribosomal mutations and the effects of these mutations on the translation process, then sorting the information in a way to curate a database that is easily accessible to researchers. Our team is divided into two subgroups: one subgroup is focusing on the effect of ribosomal mutations on cellular mechanisms of antibiotic resistance. Our subgroup is focused on identifying mutations to the ribosome and other translation factors then identifying what affect these mutations have on the translation process. Our hope is that being able to quickly access the effects of these mutations will lead to new and critical studies that will not only further C-GEM’s research and mission but will also benefit ribosomal research efforts everywhere.
Key words
Ribosome: Molecular Machine made up of approximately 60% ribosomal RNA and approximately 40% protein. They function to synthesize proteins for packaging and transport within the cell and out of the cell.
RNA: Short for ribonucleic acid. The nucleic acid that is used in key metabolic processes for all steps of protein synthesis in all living cells and carries the genetic information of many viruses.
mRNA: Messenger RNA (mRNA) carries the genetic information copied from DNA in the form of a series of three-base code “words,” each of which specifies a particular amino acid.
rRNA: Ribosomal RNA (rRNA) associates with a set of proteins to form ribosomes. These complex structures, which physically move along an mRNA molecule, catalyze the assembly of amino acids into protein chains. They also bind tRNAs and various accessory molecules necessary for protein synthesis. Ribosomes are composed of a large and small subunit, each of which contains its own rRNA molecule or molecules.2
mRNA: Messenger RNA. A single stranded RNA that leaves the nucleus and is carried to the ribosome for the ribosome to read it and create proteins
tRNA: Transfer RNA. A molecule, composed of RNA that carries amino acids to the ribosome to be added to a chain of amino acids.
Amino Acid: An organic compound containing at least one amino group and one carboxyl group centered around a central carbon atom, the α carbon, to which a variable side chain is bound.
Prokaryote: A cell that is distinguishable from a eukaryote by its lack of nucleus. DNA floats freely in the cell’s cytoplasm usually in a circular shape, sometimes referred to as plasmids.
Monomer: a molecule of any compound, mostly organic, that can react with other molecules to form very large molecules, or polymers. The essential feature of a monomer is their ability to perform multiple functions and the capacity to form chemical bonds to at least two other monomer molecules. Bifunctional monomers can form only linear, chainlike polymers, but monomers of higher functionality yield cross-linked, network polymeric products.3
Polymer: A group of monomers joined together.
E. coli: The bacterium Escherichia coli (E. coli for short) is crucial in modern biotechnology. Scientists use it to store DNA sequences from other organisms, to produce proteins and to test protein function.4
50S subunit: In prokaryotes, large subunit containing 23Sribosomal RNA (rRNA), 5S rRNA and Proteins L1-L77.30S Subunit: In prokaryotes, smaller ribosomal subunit containing 16S rRNA and proteins S1-S22
Background/Knowledge Gap
The ribosome is the protein-synthesizing structure of the cell that is composed of nucleic acid and protein in two subunits: the large 50S subunit and the smaller 30S subunit. In a complex process known as translation, the ribosome builds a polypeptide chain by adding amino acids one by one, following a sequence determined by the mRNA strand. Translation can be broken down into three main steps: initiation, elongation and termination. Elongation can be further subdivided into three additional parts including decoding, elongation and peptidyl transferase. Elongation involves elongation factors such as EF-Tu and EF-G. EF-Tu (elongation factor thermal unstable) is a protein that catalyzes the binding of the aminoacyl tRNA to the A site. EF-G (elongation factor G) works as a catalyst in prokaryotes that moves mRNA and tRNA to the ribosome. Mutations in the ribosome’s RNA and protein structures can lead to variations in the rRNA or other factors involved in the elongation process and can ultimately affect part of or all of the translation process. Our subgroup focuses on studies published on mutations of the ribosome, including the peptidyl transferase center (PTC) of the large ribosomal subunit and all supporting parts of the unit.5 We aim to collect information about PTC mutations to support future studies and further ribosomal engineering research efforts.6
Methods
Our interest in the ribosome was fueled by the wide variety of papers on ribosomal mutations dating back to the early 1970’s. We narrowed the scope of the papers that would be included in the database to papers that involved mutations to the ribosome’s RNA or proteins directly. The research papers were then refined into categories including organism the experiment was performed on, what molecule/s were mutated, which residues were mutated, what the original residue was, what the mutated residue was, what the result of the mutation. Due to the wide date range of the papers, we also included the types of experiments performed, as researchers would understand the limits of the experiments and could decide if new technology could potentially lead to a deeper understanding of the experiment.
Results
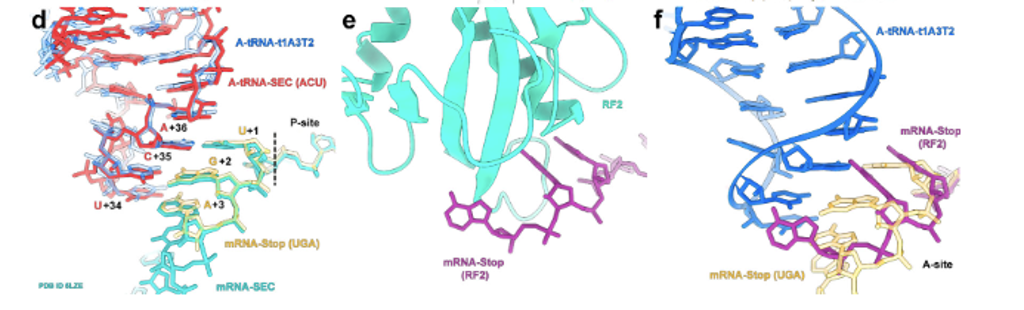
While reading peptidyl transferase mutation papers, one of the themes we followed focused on the effects of some ribosomal mutations on translation termination. Termination is one of the final steps in the process of translation and occurs when a stop codon has been reached, thus-with the help of some other release factors-resulting in the release of the peptide chain. The ribosome, just like any machinery, needs to be able to successfully terminate at the end of every process, as this is important to the overall production, function, and even structure of proteins. Fig. 2 (adapted from reference 11) is a visual representation of the interaction of the stop codon on the mRNA and with RF-2.11 Some of the papers observed how mutations to the RNA of ribosomal subunits impact this process directly or indirectly by affecting release factors that play a role in translation termination.5 Examples of mutations that affect termination include base change mutations that cause readthrough, and mutations that affect release factor 2, which is responsible for hydrolysis of peptidyl-tRNA during termination.6 Having a fundamental understanding of translation termination is essential to greater efforts in C-GEM like incorporating non-canonical amino acids and creating fully functional protein hybrids.
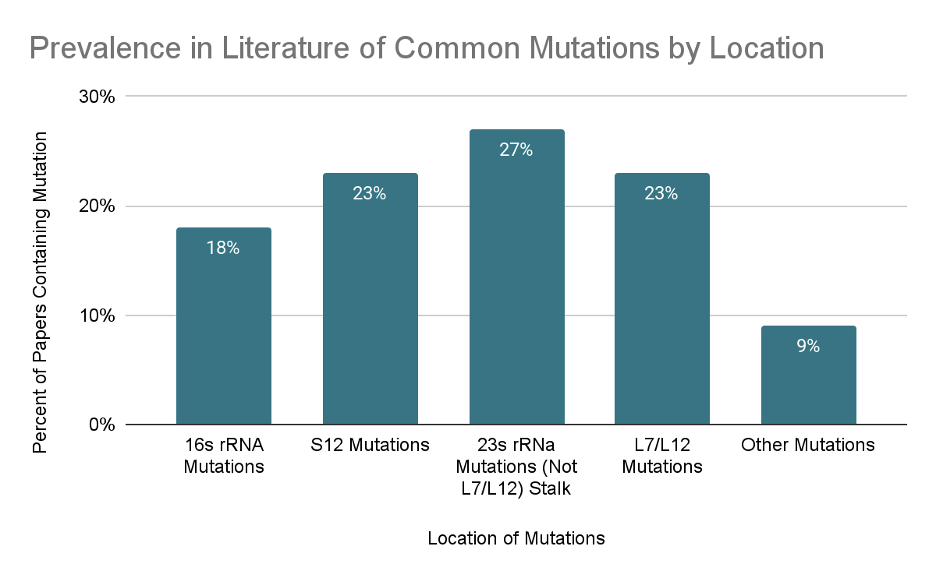
Ribosomal mutations that do not affect antibiotic resistance may affect other mechanisms of translation, including initiation of translation, rate and fidelity of translation, and termination of translation. All of these factors can potentially affect the synthesis of any engineered monomers through the promotion or inhibition of any of the mechanisms involved in translation. In addition to the expected mutations to the rRNA of the 50S and 30S subunits, a common theme among the papers we have read include mutations to the S12 protein in the small 30S ribosomal subunit. (Fig. 3) These mutations are found to be important in the inhibition or promotion of protein synthesis. This may be due to the central location of the S12 protein within the 30S ribosomal subunit’s A site, the interactions it has with the 16S rRNA backbone and the large 50S subunit.7 Other commonly studied points of mutations were found to the rRNA on the large 50S subunit as well as to the 23S rRNA of the large subunit and the 16S rRNA of the small subunit. One particular mutation that has been frequently studied is the L7/L12 stalk of the 23S rRNA which encompasses the proteins of L10 and L11 and is a hook like structure projecting from the from the anterior portion of the superior anterior region of the larger 50S subunit (Fig. 4).


Discussion
The creation of an easy-to-use database that can be updated and accessed by researchers is something that will greatly increase the efficiency of the research field in experimental design and implementation. Previous databases that have sought to quantify mutations, are no longer searchable and have not been updated since the late 1990s.12 We hope to integrate the work of these previous databases with our work to make a comprehensive database of mutations. Such a database will assist the ribosome engineering field. It is our hope that the database will continue to be maintained and updated with the help of future researchers, undergraduate students and volunteers.
References
- Research, Center for Genetically Encoded Materials. Center for Genetically Encoded Materials. Accessed July 16, 2021. https://gem-net.net/research/
- Lodish H, Berk A, Zipursky SL, et al. Molecular Cell Biology. 4th edition. New York: W. H. Freeman; 2000. Section 4.4, The Three Roles of RNA in Protein Synthesis. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21603/
- Britannica, T. Editors of Encyclopedia (2020, March 5). Monomer. Encyclopedia Britannica. https://www.britannica.com/science/monomer
- Wassenaar, T. M. (2016). Insights from 100 years of research with probiotic E. coli, European Journal of Microbiology and Immunology EuJMI, 6(3), 147-161. Retrieved Jul 16, 2021, from https://akjournals.com/view/journals/1886/6/3/article-p147.xml
- Arkov, A. L.; Freistroffer, D. V.; Ehrenberg, M.; Murgola, E. J. Mutations in RNAs of Both Ribosomal Subunits Cause Defects in Translation Termination. EMBO J. 1998, 17 (5), 1507–1514. https://doi.org/10.1093/emboj/17.5.1507
- Albers, S.; Beckert, B.; Matthies, M. C.; Mandava, C. S.; Schuster, R.; Seuring, C.; Riedner, M.; Sanyal, S.; Torda, A. E.; Wilson, D. N.; Ignatova, Z. Repurposing TRNAs for Nonsense Suppression. Nat. Commun. 2021, 12 (1), 3850. https://doi.org/10.1038/s41467-021-24076-x.
- Demirci H, Wang L, Murphy FV 4th, et al. The central role of protein S12 in organizing the structure of the decoding site of the ribosome. RNA. 2013;19(12):1791-1801. https://doi.org/10.1261/rna.040030.113
- Jahagirdar D, Jha V, Basu K, Gomez-Blanco J, Vargas J, Ortega J. Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy. RNA. 2020;26(12):2017-2030. https://doi.org/10.1261/rna.075846.120
- Schmeing, T. M., Voorhees, R. M., Kelley, A. C., Gao, Y.-G., Murphy, F. V., Weir, J. R., & Ramakrishnan, V. (2009). The Crystal Structure of the Ribosome Bound to EF-Tu and Aminoacyl-tRNA. Science, 326(5953), 688. https://doi.org/10.1126/science.1179700
- Vanja Stojković, Alexander G Myasnikov, Iris D Young, Adam Frost, James S Fraser, Danica Galonić Fujimori, Assessment of the nucleotide modifications in the high-resolution cryo-electron microscopy structure of the Escherichia coli 50S subunit, Nucleic Acids Research, Volume 48, Issue 5, 18 March 2020, Pages 2723–2732, https://doi.org/10.1093/nar/gkaa037
- Albers, S., Beckert, B., Matthies, M.C. et al. Repurposing tRNAs for nonsense suppression. Nat Commun 12, 3850 (2021). https://doi.org/10.1038/s41467-021-24076-x
- Triman Mutation Database GA Tech. https://crw-site.chemistry.gatech.edu/SAE/2B/RNA_Mutations_Triman_Database.pdf