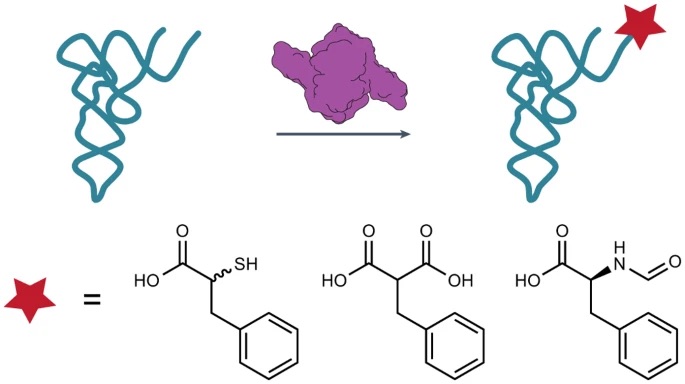
Aminoacyl-tRNA synthetases (aaRSs) catalyze the condensation of α-amino acid monomers with tRNA. The resulting aminoacyl-tRNAs are used as substrates by the ribosome to generate natural sequence-defined bio-polymers. The ribosomal synthesis of non-natural sequence-defined bio-polymers, a major C-GEM goal, demands aaRS enzymes for monomers that are not α-amino or α-hydroxy acids.
This paper describes several such enzymes, including those that do not react appreciably with natural α-amino acids. In particular, enzymes related to PylRS, the aminoacyl-tRNA synthetase that naturally incorporates pyrrolysine into proteins in certain bacteria and archaea, also accept α-thio acids, substituted malonic acids (α-carboxyl acids), and N-formyl-α-amino acids. The authors report that all of these novel monomers successfully initiated translation in vitro and monomers with reactive nucleophiles could be introduced into proteins in vivo. The high-resolution x-ray structure of one PylRS variant in complex with a substituted malonic acid substrate provides clear guidance on how to further engineer PylRS enzymes for additional novel substrates.
This work involved contributions from the Schepartz lab, Chatterjee lab, Managing Director Smaga, and Amgen Process Development.
Read the paper: https://www.nature.com/articles/s41557-023-01224-y
Read coverage of this paper in Berkeley News, Nature News