Margaret Adenusi (left) and Alyssa Virola-Iarussi (right) are undergraduate students in the 2021 C-GEM summer research program, who are analyzing acylation data to identify structure-activity relationships that will enable efficient flexizyme-mediated tRNA misacylation with backbone-altered monomers.
Abstract
Presume a world where pharmaceutical drugs do not expire, and we can have at-home treatments readily accessible. An environment where we can have cleaner water and recycling becomes the standard. Envision a world where ribosomes that natively utilize 20 L-alpha-amino acids are repurposed to create wholly unnatural chemical polymers. As part of the Center for Genetically Encoded Materials (C-GEM), our overall goal is to synthesize such chemically diverse polymers. These unnatural polymers will pave the way for new drug development and therapeutic remedies. Researchers are currently working on a way to alter “nature’s protein-synthesizing machine”, known as the ribosome. Specifically, our team is investigating the use of flexizymes in translation to charge diverse unnatural monomers onto tRNAs.
Key words
tRNA: (transfer) ribonucleic acid which delivers amino acids to the ribosome active site.
Ribosome: Macromolecular machines that are responsible for linking amino acids together, forming protein polymers in the process of translation
Microhelix: Small RNA used as a tool to mimic tRNA in assays to measure aminoacylation
In vitro vs in vivo: Experiment conducted outside a living organism vs inside a living organism
Aminoacylation: The attachment of an amino acid to a tRNA
Aminoacyl-tRNA synthetases: (also known as tRNA-ligase) An enzyme responsible for attaching the amino acid to its corresponding tRNA
Non-canonical amino acids: Amino acids that are not found within the main 20 amino acids
Flexizyme: A highly flexible RNA-based enzyme that attaches monomers onto tRNA
Introduction
C-GEM is interested in the structure and function of the ribosome and its translation factors. The organization’s main goal is to repurpose “nature’s protein-synthesizing machine,” or ribosomes, in an effort to create unnatural polymers made of monomers such as beta and gamma amino acids. The use of unnatural polymers will pave the way for new therapeutic remedies, which have been a mutual interest for our team. Throughout this summer, working as a member of C-GEM has inspired us to pursue careers in the sciences. We have gained experience working within a team, networked with commendable scientists, and gained research experience virtually. The knowledge we are gaining does not stop with learning about the ribosome. Our team’s focus has been obtaining information on various monomers that have been mis-acylated through the use of flexizymes.
Our interest in flexizymes derives from their ability to charge various monomers onto tRNAs during aminoacylation. Compared to typical aminoacyl-tRNA synthetases (aaRS), flexizymes do not possess the limitation of only accepting one specific amino acid to be charged. In other words, flexizymes can accept virtually any amino acid as an acyl-donor, as long as the flexizyme can recognize the leaving group of the substrate.1 This relieves the bottleneck of tRNA aminoacylation and allows us to assess if the monomers are valuable ribosomal substrates. As a result, the process of developing unnatural polymers through ribosomal translation is accelerated.
Even though the use of flexizymes is promising for developing unnatural polymers, they only function in vitro. In addition, the turnover rate for this RNA enzyme is low. Consequently, the flexizyme reaction time is slower compared to reactions using aaRS. Our work on compiling data on various unnatural monomers into a shared database will provide future researchers with readily accessible data regarding flexizymes, their substrates, and key information about reaction conditions and yields.
Methods
In order to compiling information on various flexizyme reactions, we read a lot of scientific literature, beginning with “Flexizymes: Their Evolutionary History and the Origin of Catalytic Function”.2 This review paper provided comprehensive information on the history of flexizymes and their role in translation,2 and enabled us to find related articles in which researchers experimented on the use of flexizymes with diverse substrates. Together we found and read ten articles and used Zotero to collect, organize, and document all our papers.
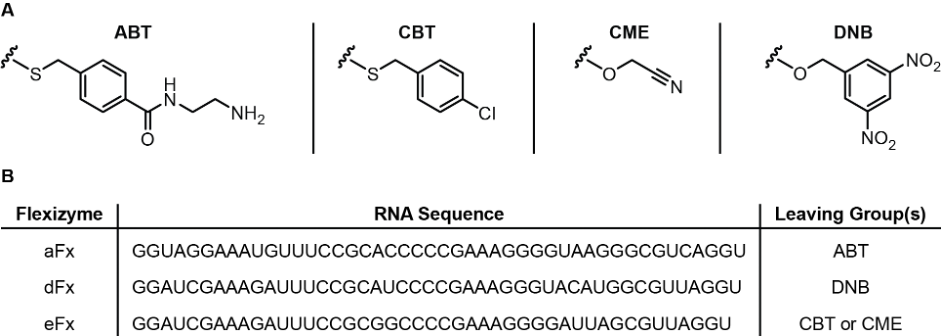
After reading a few papers, we compiled information we thought would be useful to provide to future researchers into a google spreadsheet such as gamma-amino acids and benzoic acid derivatives. Each monomer had a CME, DBE, or ABT leaving grouping attached to its substrate and a corresponding flexizyme that recognized that leaving group. Figure 1 shows the relationship between each flexizyme and the leaving group. To help organize the various monomers created, the names were recorded and their structures were drawn using a SMILES (Simplified Molecular Input Line Entry System) notation generator.12 SMILES was an easily learned and flexible notation that made inputting structures convenient and unambiguous. Lastly, information about the reaction times and acylation efficiency for microhelix RNA were recorded.
The data spreadsheet also included the type of flexizyme, type of tRNA used, type of ribosome, and in vitro translation protocol. Gathering information on these components was done by observing the figures and the figure captions of each paper. In-depth information on the substrate, the monomer used, percent yield, tRNA, and flexizyme sequences were obtained a paper’s supplementary information.
Eventually, the data in this spreadsheet can be imported into a more formal database that can be shared with C-GEM researchers and the public. In addition to improving the accessibility of existing flexizyme research, the database will be updated as new flexizyme-compatible monomers are reported in the literature.
Results and Discussion
During the process of filling out the database, the major connections we found between the papers we read were how well flexizymes charge artificial monomers onto tRNA. This project helped point out that monomer identity and microhelix conditions play an important role in how well the flexizyme can catalyze a reaction. Furthermore, we noticed that monomers seemed to produce a higher microhelix yield when the reaction ran for 120 hours versus 16 hours. This supports the claim that the flexizyme takes more time to catalyze the reaction versus the standard aaRS. This may also suggest that researchers who want to improve misacylation yield should allow the reaction to proceed as long as possible. Moreover, it appears that reactions performed at a pH of 8.0 produced a higher yield than those under a pH of 7.5.
Overall, this virtual experience was challenging. Although reading papers on a weekly basis was a heavy load, it was fulfilling to know that our work was beneficial to the broader C-GEM goals and community. We hope that further work to expand and use this database will help researchers make more informed decisions when using flexizyme to acylate tRNAs with unnatural monomers. Even though there is much left to accomplish, we believe that our project and C-GEM as a whole is on an exciting path investigating “nature’s-protein-synthesizing machine.” It is truly commendable that an organization can develop a project that will serve as a benefit to society for the time to come, making science a never-ending journey for new discoveries.
References
- Morimoto, J. et al. Flexizymes: Their Evolutionary History and the Origin of Catalytic Function. Accounts of Chemical Research, 2011. 44 (12), 1359–1368. https://pubs.acs.org/doi/10.1021/ar2000953
- Ohshiro, Y. et al. Ribosomal Synthesis of Backbone-Macrocyclic Peptides Containing γ-Amino Acids. ChemBioChem 2011, 12 (8), 1183–1187. https://doi.org/10.1002/cbic.201100104
