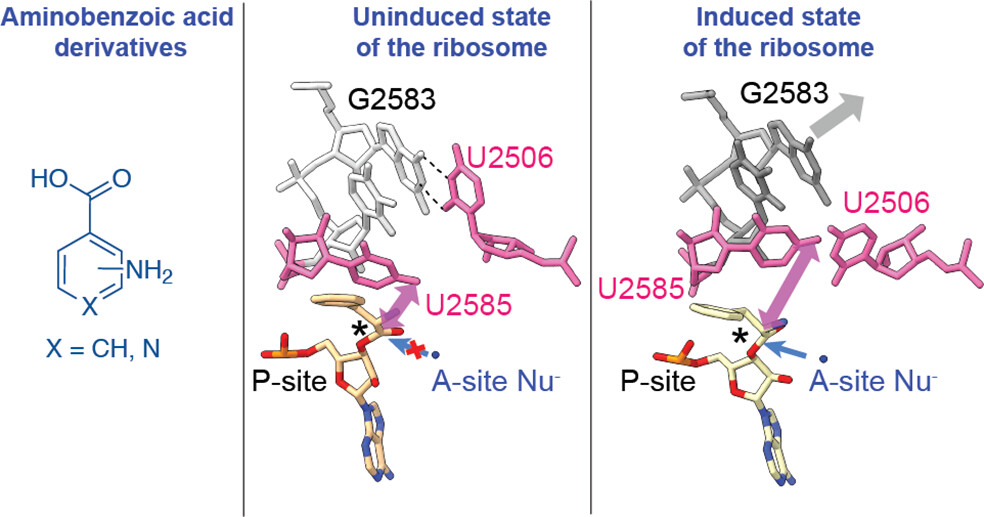
Although the E. coli ribosome can incorporate a variety of non-L-α-amino acid monomers into polypeptide chains in vitro, there exists no high-resolution structural information regarding their positioning within the catalytic center of the ribosome, the peptidyl transferase center (PTC). Thus, details regarding the mechanism of amide bond formation and the structural basis for differences and defects in incorporation efficiency remain unknown.
This paper reports high-resolution cryo-EM structures of the ribosome with three aminobenzoic acid derivatives charged on tRNA bound in the aminoacyl-tRNA site (A-site). The structures reveal that aromatic ring of each monomer sterically blocks the ribosome from adopting its “induced fit” required for catalysis. Structures also reveal disruptions to the bound water network that is believed to facilitate formation and breakdown of the tetrahedral intermediate. Together, the cryo-EM structures reported here provide a mechanistic rationale for differences in reactivity of aminobenzoic acid derivatives relative to L-α-amino acids and identify stereochemical constraints on the size and geometry of non-natural monomers that can be accepted efficiently by wild-type ribosomes.
This work was a collaboration between the Schepartz and Cate groups.
Read the paper: https://pubs.acs.org/doi/10.1021/acscentsci.3c00153
Read coverage of this paper in Berkeley News, Nature News